
The Machine Learning for Medical Language Lab studies the use of machine learning-based natural language processing for biomedical and health-related use cases.

Our mission is to develop and implement Natural Language Processing (NLP) technologies to apply to the electronic medical record. These technologies include core NLP tasks such as relation extraction, coreference resolution, and parsing, and make use of statistical machine learning methods. In order to use many machine learning methods, manually labeled (annotated) domain- and task-specific data is required. To that end, we are heavily involved in many different clinical document annotation projects. Since manual annotation is a time-consuming, painstaking, expensive process, it is also our goal to develop and use algorithms that minimize the required amount of labeled data required while maximizing the use of existing labeled data.

The Majumder Lab specializes in artificial intelligence & machine learning (AI/ML) methods for public health problems. Among others, current problem areas include emerging & vaccine-preventable infections; medical misinformation; and outcome disparities in marginalized populations.

The Predictive Medicine Group works to develop novel approaches for predicting human health. Our diverse group of researchers, clinicians, mathematicians, computer scientists and biologists develop advanced predictive models for a wide range of applicants, including disease risk prediction, predictive pharmacovigilance, predictive health system dynamics and real-time public health surveillance.

SMART Health IT advances medicine, discovery and public health through parsimonious, open standards, application programming interfaces, laws and regulation, and is best known for the SMART on FHIR API.

The Computational Epidemiology Lab conducts a diverse set of projects to predict patterns of disease, analyze patterns of epidemics, use of satellite data and public health surveillance tools.
Our lab aims to decode the key principles of cancer genomics in tumor development and use them as an “Achilles heel” for targeted cancer therapies. We are a diverse group of scientists with a common research interest in cancer genomics, tumor biology, and targeted therapies. We combine statistics, machine learning, tumor biology, and functional genomics for a data-driven understanding of the molecular architecture of tumor genomes and the development of genome-inspired therapies. We are committed to training a new generation of cancer researchers from computational, experimental, and clinical backgrounds who share our core values of teaching and mentorship, collegiality and collaboration, inclusion and diversity, and mutual respect and open communication.

The Translational Omics Medicine Lab develops methods to molecularly characterize patients for research and discovery.

We are a multi-disciplinary group, open to students in clinical and biomedical sciences, computer science, statistics, engineering, and related fields. We seek clarity and fairness from complex problems that necessitate interdisciplinary approaches. Our goals are to cultivate an environment for critical thinking, research creativity and open scientific collaboration. In addition, this lab is a good fit for those who wish to gain expertise in biomedical informatics and machine learning, especially when interpretability and fairness are central concerns.

The RIR&D Group is focused on the development and application of innovative, open-source, data-driven solutions to challenges faced by longitudinal disease registries and related studies. A principal focus of our group is informatics-based strategies to enhance the collection and sharing of clinical research data within the 75+ site Childhood Arthritis and Rheumatology Research Alliance (CARRA) Registry, serving as a paradigm for other similar longitudinal chronic disease registries. Members of the CHIP RIR&D Group include specialists in software architecture and development for multi-site/multi-source regulatory-grade clinical research data warehousing, research data coordination, research analytics and computable phenotyping, and related IRB and regulatory/legal matters for clinical trials.

The Manrai Lab is a team of machine learning scientists, clinicians, and biomedical data scientists working to improve medical decision making by developing computational approaches that incorporate rich and deep representations of clinical state and an individual's identity into care. Active projects include: improving genetic variant classification and quantifying risk ("penetrance") for inherited heart disease, measuring "normal" variation for blood lab biomarkers across populations focused on creatinine and kidney disease, developing semi-supervised learning approaches for multi-modal imaging, and modeling reproducibility in integrative biomedical studies using meta-science ("science of science") approaches.

The Pediatric Therapeutics and Regulatory Science Initiative provides a forum for collaboration with academia, patients, policymakers, and industry. Its aim is to advance the development and evidence-based use of novel therapeutics for children globally.

The LNN develops computational methods for extracting digital biomarkers for brain disorders from EEG measurements. Our approach to EEG signal analysis is based in complex dynamical systems theory. Entropy measures, recurrence plot and recurrence network analysis, neural synchronization, and tensor data structures all play a role.
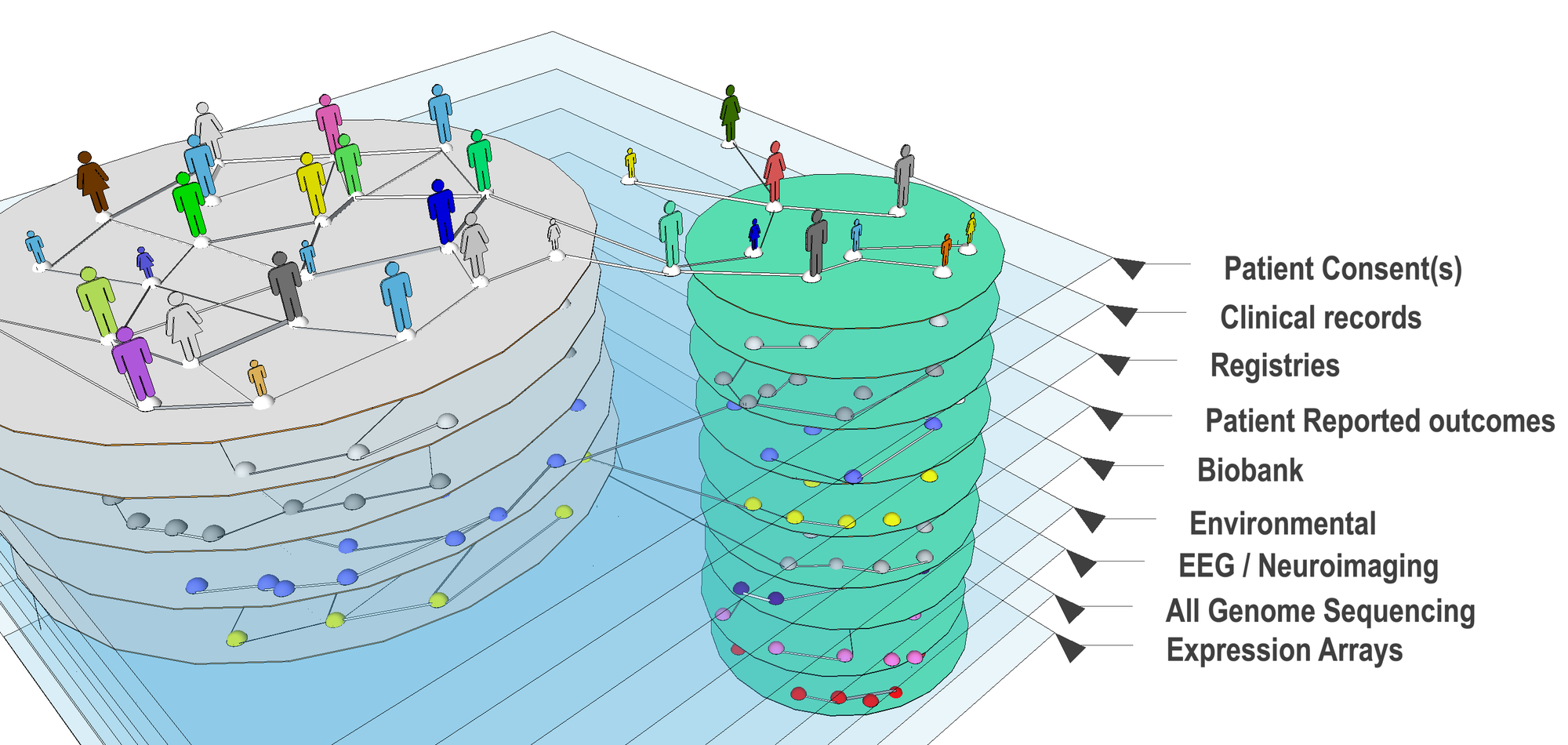
The Avillach Lab investigates translational bioinformatics, specifically in integrating multiple heterogeneous sources of clinical and genomics data in a meaningful way.

The Strober Lab is a computational research group focused on developing statistical and machine learning methods applied to human genetic and genomic data. Our goals are to elucidate disease biology and advance genomically-informed precision medicine. We are particularly interested in modeling the regulatory cascade from DNA through molecular phenotypes, such as transcript and protein levels, to diseases. We also aim to integrate multimodal omics with the rich clinical data to refine diagnoses and guide precision-medicine interventions.




